Section 13: data.table
Admin
Announcements
- Today is our last section!
- Office hours at usual time/place on Friday.
- Whoever submits your problem set on bCourses has access to my comments. The solutions are also available for problem sets 1–3 (and 4 will be up shortly).
- From problem set 3: Remember when you use the delta method that you need to take derivatives with respect to all of parameters in model. In the case of the problem in the problem set, each of the derivatives was equal to zero except one, but the derivation that many of you provided for the delta-method-based standard error of our estimator was no quite right.
- Course evaluations are available!
Last section
In our previous section, we discussed spatial—focusing on one of the two major classes of spatial data: vector data, which include polygons, lines, and points.
This week
Spatial data
Today we will wrap up our discussion of spatial data, covering three topics:
- Raster data: our remaining major class of spatial data
- Geocoding: moving from addresses to geographic coordinates
- Leaflet: an awesome tools for creating interactive maps
See Section 12b for this completion of spatial data.
data.table
In the second portion of this section, we will R’s data.table package and its uses for cleaning/prepping datasets. data.table is particularly useful for large datasets, so we will use it on a fairly large dataset.
What you will need
Packages:
- New:
data.table - Old: magrittr`
data.table
R’s data.table package provides methods for fast (potentially grouped) manipulation of large datasets—aggregation, joins, modifications—without copying the dataset repeatedly. The syntax is not quite as clear as the “verbs” used by dplyr and the rest of the hadleyverse, but the efficiency benefits are substantial when you have large datsets.
R setup
Let’s get started. First, set up R.
# General R setup ----
# Options
options(stringsAsFactors = F)
# Load packages
pacman::p_load(data.table, magrittr)
# Add an option for data.table
options(datatable.print.nrows = 20)The option options(datatable.print.nrows = 20) tells R that we do not want to print the whole data table if it has more than 20 rows—a handy feature that prevents you from printing a billion rows to screen.1
Creating a data table
Alright, let’s create a data table.2 To create a data table—or to convert an existing data frame or matrix to a data table—you use the data.table() function just as you would use the data.frame() function. Let’s create a simple data table with 30 rows and four columns named a, b, c, and d. Note: I’m going to generally use the suffix _dt to denote data tables in our R scripts.
# Set the seed
set.seed(12345)
# Create our data table
our_dt <- data.table(
a = 101:130,
b = rnorm(30),
c = runif(30),
d = rep(1:5))So what is this data table object that we just created?
# Check the class
our_dt %>% class()## [1] "data.table" "data.frame"# Check the dimensions, length, and names
our_dt %>% dim()## [1] 30 4our_dt %>% length()## [1] 4our_dt %>% names()## [1] "a" "b" "c" "d"This data table is still a data.frame, but it is also a data.table—similar to how we previously created objects that simultaneously had the classes data.frame, tbl_df, and tbl. The dimensions, length, and names are exactly what we expected them to be. Good.
Let’s print the data table to screen.
# Print our_dt to screen
our_dt## a b c d
## 1: 101 0.5855288 0.79156780 1
## 2: 102 0.7094660 0.25868432 2
## 3: 103 -0.1093033 0.98598383 3
## 4: 104 -0.4534972 0.75687374 4
## 5: 105 0.6058875 0.97977825 5
## ---
## 26: 126 1.8050975 0.30503175 1
## 27: 127 -0.4816474 0.82565686 2
## 28: 128 0.6203798 0.50234464 3
## 29: 129 0.6121235 0.80357263 4
## 30: 130 -0.1623110 0.06063998 5I like this feature of data tables: you get to see the first and last five rows for each variable.
General syntax
The general syntax for data tables is slightly different from what we’ve seen so far. The main idea for the syntax of a data table is DT[i, j, by], where DT is our data table, i references the rows that we want, j references the columns, and by provides the variables with which we want group our data. If you want all of the rows, simply omit i (but keep the comma). Likewise for j. And if you do not want to group your data, then omit by (you do not need to include the comma between j and by if you do not use the by argument).
Indexing columns
Let’s talk about indexing columns/variables.
Accessing a column
You can still use the $ to access columns, but this practice is discouraged because it is less efficient and generalizable than data.table’s other methods for accessing columns.
So how do you access columns? You define j as either the variable names or numbers. If we want to grab b (the second variable), we can define j as b or "b" or 2. However, some of these methods will result in vector of the values in the b column, while others will result in a data table with only the column b. Finally, we can treat a data table (or data frame) as a list and reference a column with double square brackets and the column’s name (e.g., our_dt[["b"]]) or number.
Six options for grabbing the b variable:
our_dt$b## [1] 0.5855288 0.7094660 -0.1093033 -0.4534972 0.6058875 -1.8179560
## [7] 0.6300986 -0.2761841 -0.2841597 -0.9193220 -0.1162478 1.8173120
## [13] 0.3706279 0.5202165 -0.7505320 0.8168998 -0.8863575 -0.3315776
## [19] 1.1207127 0.2987237 0.7796219 1.4557851 -0.6443284 -1.5531374
## [25] -1.5977095 1.8050975 -0.4816474 0.6203798 0.6121235 -0.1623110our_dt[, b]## [1] 0.5855288 0.7094660 -0.1093033 -0.4534972 0.6058875 -1.8179560
## [7] 0.6300986 -0.2761841 -0.2841597 -0.9193220 -0.1162478 1.8173120
## [13] 0.3706279 0.5202165 -0.7505320 0.8168998 -0.8863575 -0.3315776
## [19] 1.1207127 0.2987237 0.7796219 1.4557851 -0.6443284 -1.5531374
## [25] -1.5977095 1.8050975 -0.4816474 0.6203798 0.6121235 -0.1623110our_dt[, "b"]## b
## 1: 0.5855288
## 2: 0.7094660
## 3: -0.1093033
## 4: -0.4534972
## 5: 0.6058875
## ---
## 26: 1.8050975
## 27: -0.4816474
## 28: 0.6203798
## 29: 0.6121235
## 30: -0.1623110our_dt[, 2]## b
## 1: 0.5855288
## 2: 0.7094660
## 3: -0.1093033
## 4: -0.4534972
## 5: 0.6058875
## ---
## 26: 1.8050975
## 27: -0.4816474
## 28: 0.6203798
## 29: 0.6121235
## 30: -0.1623110our_dt[["b"]]## [1] 0.5855288 0.7094660 -0.1093033 -0.4534972 0.6058875 -1.8179560
## [7] 0.6300986 -0.2761841 -0.2841597 -0.9193220 -0.1162478 1.8173120
## [13] 0.3706279 0.5202165 -0.7505320 0.8168998 -0.8863575 -0.3315776
## [19] 1.1207127 0.2987237 0.7796219 1.4557851 -0.6443284 -1.5531374
## [25] -1.5977095 1.8050975 -0.4816474 0.6203798 0.6121235 -0.1623110our_dt[[2]]## [1] 0.5855288 0.7094660 -0.1093033 -0.4534972 0.6058875 -1.8179560
## [7] 0.6300986 -0.2761841 -0.2841597 -0.9193220 -0.1162478 1.8173120
## [13] 0.3706279 0.5202165 -0.7505320 0.8168998 -0.8863575 -0.3315776
## [19] 1.1207127 0.2987237 0.7796219 1.4557851 -0.6443284 -1.5531374
## [25] -1.5977095 1.8050975 -0.4816474 0.6203798 0.6121235 -0.1623110Finally, imagine inside of a function, you have an object named my_col, which references "b", and you want to grab the column referenced by my_col. You cannot grab the column b using our_dt[,my_col], because data.table will think you are referencing a column named my_col rather than looking at the value assigned to my_col. To tell data.table to look at the value inside of my_col, you have three options:
- Use the list-style syntax:
our_dt[[my_col]] - Add the argument
with = Fto the end of your indexing:our_dt[, .my_col, with = F] - Use double periods in front of
my_col:our_dt[, ..my_col]
Again, which option you choose affects the type of object that data.table returns.
Let’s see these solutions in action:
# Define 'my_col' as "b"
my_col <- "b"
# The erroneous use
our_dt[, my_col]## Error in `[.data.table`(our_dt, , my_col): j (the 2nd argument inside [...]) is a single symbol but column name 'my_col' is not found. Perhaps you intended DT[,..my_col] or DT[,my_col,with=FALSE]. This difference to data.frame is deliberate and explained in FAQ 1.1.# The list use
our_dt[[my_col]]## [1] 0.5855288 0.7094660 -0.1093033 -0.4534972 0.6058875 -1.8179560
## [7] 0.6300986 -0.2761841 -0.2841597 -0.9193220 -0.1162478 1.8173120
## [13] 0.3706279 0.5202165 -0.7505320 0.8168998 -0.8863575 -0.3315776
## [19] 1.1207127 0.2987237 0.7796219 1.4557851 -0.6443284 -1.5531374
## [25] -1.5977095 1.8050975 -0.4816474 0.6203798 0.6121235 -0.1623110# The 'with = F' option
our_dt[, my_col, with = F]## b
## 1: 0.5855288
## 2: 0.7094660
## 3: -0.1093033
## 4: -0.4534972
## 5: 0.6058875
## ---
## 26: 1.8050975
## 27: -0.4816474
## 28: 0.6203798
## 29: 0.6121235
## 30: -0.1623110# The double-period option
our_dt[, ..my_col]## b
## 1: 0.5855288
## 2: 0.7094660
## 3: -0.1093033
## 4: -0.4534972
## 5: 0.6058875
## ---
## 26: 1.8050975
## 27: -0.4816474
## 28: 0.6203798
## 29: 0.6121235
## 30: -0.1623110Accessing multiple columns
The [i,j,by] syntax still applies for grabbing multiple columns, as does the with = F option. The big change here is that you do not want to use the c() function in conjunction with unquoted column names. Instead, use list() or simply .(). Let’s grab the first two columns—a and b.
# Using 'list'
our_dt[, list(a, b)]## a b
## 1: 101 0.5855288
## 2: 102 0.7094660
## 3: 103 -0.1093033
## 4: 104 -0.4534972
## 5: 105 0.6058875
## ---
## 26: 126 1.8050975
## 27: 127 -0.4816474
## 28: 128 0.6203798
## 29: 129 0.6121235
## 30: 130 -0.1623110# Using the period
our_dt[, .(a, b)]## a b
## 1: 101 0.5855288
## 2: 102 0.7094660
## 3: 103 -0.1093033
## 4: 104 -0.4534972
## 5: 105 0.6058875
## ---
## 26: 126 1.8050975
## 27: 127 -0.4816474
## 28: 128 0.6203798
## 29: 129 0.6121235
## 30: 130 -0.1623110What about column numbers?
# List with numbers? No.
our_dt[, list(1, 2)]## V1 V2
## 1: 1 2# c() with numbers and 'with = F'? Yes.
our_dt[, c(1, 2), with = F]## a b
## 1: 101 0.5855288
## 2: 102 0.7094660
## 3: 103 -0.1093033
## 4: 104 -0.4534972
## 5: 105 0.6058875
## ---
## 26: 126 1.8050975
## 27: 127 -0.4816474
## 28: 128 0.6203798
## 29: 129 0.6121235
## 30: 130 -0.1623110Again, if you want reference columns via another object, you need to use with = F.
# Define the object
my_cols <- c("a", "b")
# Access the columns using the object
our_dt[, my_cols, with = F]## a b
## 1: 101 0.5855288
## 2: 102 0.7094660
## 3: 103 -0.1093033
## 4: 104 -0.4534972
## 5: 105 0.6058875
## ---
## 26: 126 1.8050975
## 27: 127 -0.4816474
## 28: 128 0.6203798
## 29: 129 0.6121235
## 30: 130 -0.1623110Indexing rows
If you want to simply grab rows by their number, you can do so just as you would with an ordinary data frame:
# Grab some rows
our_dt[c(1, 5, 7, 10), ]## a b c d
## 1: 101 0.5855288 0.7915678 1
## 2: 105 0.6058875 0.9797782 5
## 3: 107 0.6300986 0.9487072 2
## 4: 110 -0.9193220 0.9464308 5You can begin to see the power of data.table and its notation when you want to select rows using some criteria, as you would do with filter() in dplyr. Let’s grab values of b below 0 and values of c above 0.5.
# Grab b < 0 and c > 0.5
our_dt[(b < 0) & (c > 0.5),]## a b c d
## 1: 103 -0.1093033 0.9859838 3
## 2: 104 -0.4534972 0.7568737 4
## 3: 109 -0.2841597 0.6003570 4
## 4: 110 -0.9193220 0.9464308 5
## 5: 111 -0.1162478 0.6883534 1
## 6: 117 -0.8863575 0.9614473 2
## 7: 118 -0.3315776 0.6549605 3
## 8: 127 -0.4816474 0.8256569 2The parentheses here are not necessary—I just like them for ease of reading the code.
The data.table package also provides a handy function between() that helps you filter values between a min and a max. Rather than grabbing values of b less than 0, let’s restrict our rows to values of b between -0.25 and 0.25. Finally, in addition to the rows we reference, let’s grab only columns a and c.
# Grab b in [-0.25,0.25] and c > 0.5
our_dt[between(b, -0.25, 0.25) & (c > 0.5), .(a, c)]## a c
## 1: 103 0.9859838
## 2: 111 0.6883534To sample rows from your data table, you can apply R’s base functions sample() and sample.int(). Sub-sampling3 is a very useful tool when you are writing code to clean/analyze a huge dataset. Start on small subset of your data to write and test your code, then move up to the big leagues.
To sample 10 rows from our (already small) data table:
# Set the seed
set.seed(12345)
# Sample
our_dt[sample(10),]## a b c d
## 1: 108 -0.2761841 0.1494579 3
## 2: 110 -0.9193220 0.9464308 5
## 3: 107 0.6300986 0.9487072 2
## 4: 109 -0.2841597 0.6003570 4
## 5: 103 -0.1093033 0.9859838 3
## 6: 101 0.5855288 0.7915678 1
## 7: 102 0.7094660 0.2586843 2
## 8: 104 -0.4534972 0.7568737 4
## 9: 106 -1.8179560 0.2189478 1
## 10: 105 0.6058875 0.9797782 5Adding/manipulating columns
You have two options for adding and manipulating columns in a data table.4 The first is the rather strange syntax :=; the second is the function set().
Let’s start with := route for creating and manipulating variables. Recall the i,j notation we discussed above. As j denotes columns, we will create and manipulate columns in the j place of our data table. Perhaps more clearly, if we want to create a column of ones called ones in our data table, we write:
# Define a column of ones
our_dt[, ones := 1]# Check our data frame
our_dt## a b c d ones
## 1: 101 0.5855288 0.79156780 1 1
## 2: 102 0.7094660 0.25868432 2 1
## 3: 103 -0.1093033 0.98598383 3 1
## 4: 104 -0.4534972 0.75687374 4 1
## 5: 105 0.6058875 0.97977825 5 1
## ---
## 26: 126 1.8050975 0.30503175 1 1
## 27: 127 -0.4816474 0.82565686 2 1
## 28: 128 0.6203798 0.50234464 3 1
## 29: 129 0.6121235 0.80357263 4 1
## 30: 130 -0.1623110 0.06063998 5 1Notice that there is no re-defining of the data table using <- or %<>%. We simple define a new column, and then the column exists. This change is part of data.table’s efficiency.
Similarly, if we want create a column named abc that is the product of the columns a, b, and c, then we write
# Define abc
our_dt[, abc := a * b * c]# Check our data frame
our_dt## a b c d ones abc
## 1: 101 0.5855288 0.79156780 1 1 46.81206
## 2: 102 0.7094660 0.25868432 2 1 18.71983
## 3: 103 -0.1093033 0.98598383 3 1 -11.10044
## 4: 104 -0.4534972 0.75687374 4 1 -35.69697
## 5: 105 0.6058875 0.97977825 5 1 62.33171
## ---
## 26: 126 1.8050975 0.30503175 1 1 69.37712
## 27: 127 -0.4816474 0.82565686 2 1 -50.50478
## 28: 128 0.6203798 0.50234464 3 1 39.89049
## 29: 129 0.6121235 0.80357263 4 1 63.45325
## 30: 130 -0.1623110 0.06063998 5 1 -1.27953You can manipulate a column in the same way that you define a column, e.g.,
# Re-define the column
our_dt[, ones := 111]# Check our data frame
our_dt## a b c d ones abc
## 1: 101 0.5855288 0.79156780 1 111 46.81206
## 2: 102 0.7094660 0.25868432 2 111 18.71983
## 3: 103 -0.1093033 0.98598383 3 111 -11.10044
## 4: 104 -0.4534972 0.75687374 4 111 -35.69697
## 5: 105 0.6058875 0.97977825 5 111 62.33171
## ---
## 26: 126 1.8050975 0.30503175 1 111 69.37712
## 27: 127 -0.4816474 0.82565686 2 111 -50.50478
## 28: 128 0.6203798 0.50234464 3 111 39.89049
## 29: 129 0.6121235 0.80357263 4 111 63.45325
## 30: 130 -0.1623110 0.06063998 5 111 -1.27953To delete a column, you redefine the column as NULL. Let’s delete the column ones.
# Delete 'ones'
our_dt[, ones := NULL]our_dt## a b c d abc
## 1: 101 0.5855288 0.79156780 1 46.81206
## 2: 102 0.7094660 0.25868432 2 18.71983
## 3: 103 -0.1093033 0.98598383 3 -11.10044
## 4: 104 -0.4534972 0.75687374 4 -35.69697
## 5: 105 0.6058875 0.97977825 5 62.33171
## ---
## 26: 126 1.8050975 0.30503175 1 69.37712
## 27: 127 -0.4816474 0.82565686 2 -50.50478
## 28: 128 0.6203798 0.50234464 3 39.89049
## 29: 129 0.6121235 0.80357263 4 63.45325
## 30: 130 -0.1623110 0.06063998 5 -1.27953If you want to define/manipulate multiple columns, you add parentheses to := and separate the new columns with commas. Let’s delete abc and at the same time add the two-way products ab and ac.
our_dt[, `:=`(
abc = NULL,
ab = a * b,
ac = a *c
)]our_dt## a b c d ab ac
## 1: 101 0.5855288 0.79156780 1 59.13841 79.948348
## 2: 102 0.7094660 0.25868432 2 72.36553 26.385800
## 3: 103 -0.1093033 0.98598383 3 -11.25824 101.556335
## 4: 104 -0.4534972 0.75687374 4 -47.16371 78.714869
## 5: 105 0.6058875 0.97977825 5 63.61818 102.876716
## ---
## 26: 126 1.8050975 0.30503175 1 227.44229 38.434001
## 27: 127 -0.4816474 0.82565686 2 -61.16922 104.858422
## 28: 128 0.6203798 0.50234464 3 79.40861 64.300114
## 29: 129 0.6121235 0.80357263 4 78.96393 103.660869
## 30: 130 -0.1623110 0.06063998 5 -21.10043 7.883198As I mentioned above, you can also define and manipulate columns using the data.table function set(). set() also uses the i,j syntax. set() takes the following arguments:
x: the name of data tablei: the row numbers which you want to receive the new values (omit for all rows)j: the name (or number) of the columnvalue: the value to assign
Let’s use set() to add back our column of ones… and then delete it.
# Add column of ones using 'set'
set(x = our_dt, j = "ones", value = 1)
# Delete column of ones using 'set'
set(x = our_dt, j = "ones", value = NULL)set() is very helpful within a function, since it allows you to easily define the rows, column name (as a character), and values.
Setting names
Continuing in the spirit of set(), data.table also provides a function to set column names. Why? Because names(our_dt) <-… is, again, an inefficient way to set the column names: it when you use <-, you are copying the entire data frame to simply change a few names.
Let’s imagine that we want to change all of our variable names to all capital letters. The function toupper() changes characters to uppercase, so we could write names(our_dt) <- toupper(names(our_dt)), but we want to be efficient, so let’s use setnames().
# Change names to uppercase
setnames(our_dt, toupper(names(our_dt)))What if we only want to change a single name? setnames() still works! It accepts the arguments old and new, so we can single out the old variable name and replace it with a new name. Let’s change "AB" to "ab".
# Change 'AB' to 'ab'
setnames(our_dt, old = "AB", new = "ab")
# Check our data table
our_dt## A B C D ab AC
## 1: 101 0.5855288 0.79156780 1 59.13841 79.948348
## 2: 102 0.7094660 0.25868432 2 72.36553 26.385800
## 3: 103 -0.1093033 0.98598383 3 -11.25824 101.556335
## 4: 104 -0.4534972 0.75687374 4 -47.16371 78.714869
## 5: 105 0.6058875 0.97977825 5 63.61818 102.876716
## ---
## 26: 126 1.8050975 0.30503175 1 227.44229 38.434001
## 27: 127 -0.4816474 0.82565686 2 -61.16922 104.858422
## 28: 128 0.6203798 0.50234464 3 79.40861 64.300114
## 29: 129 0.6121235 0.80357263 4 78.96393 103.660869
## 30: 130 -0.1623110 0.06063998 5 -21.10043 7.883198Finally, let’s change the names back to lowercase, using tolower().
# Change names back to lowercase
setnames(our_dt, tolower(names(our_dt)))Summarizing columns
data.table’s i,j,by syntax also allows you to quickly summarize variables. For instance, if you want to take the means of a and b:
# Mean of 'a' and 'b'
our_dt[, .(mean(a), mean(b))]## V1 V2
## 1: 115.5 0.07880701However, V1 and V2 are not the more informative names. With a few more keystrokes, we can add names:
# Mean of 'a' and 'b', named
our_dt[, .(mean_a = mean(a), mean_b = mean(b))]## mean_a mean_b
## 1: 115.5 0.07880701Nice, right?
What if we want to take the means of a and b, grouped by the variable d (which takes on the values 1, 2, 3, 4, and 5)? Enter the by part of i,j,by!
# Mean of 'a' and 'b' grouped by 'd'
our_dt[, .(mean_a = mean(a), mean_b = mean(b)), by = d]## d mean_a mean_b
## 1: 1 113.5 0.342157388
## 2: 2 114.5 0.540776135
## 3: 3 115.5 -0.061730962
## 4: 4 116.5 -0.006290287
## 5: 5 117.5 -0.420877223Hopefully you are starting to get on board with this data.table thing.
data.table has a few special commands that are only accessible inside of the square brackets of a data table. One of them is .N, which gives the number of observations for the given data table/group. For instance, we can access the final observation of a data table using
# Last observation
our_dt[.N,]## a b c d ab ac
## 1: 130 -0.162311 0.06063998 5 -21.10043 7.883198We can also use .N as a summary statistic to count the number of observations within each group:
our_dt[, .N, by = d]## d N
## 1: 1 6
## 2: 2 6
## 3: 3 6
## 4: 4 6
## 5: 5 6You can also add summaries back into the data table, using the same notation we used above. For instance, if we wan to add the mean of a, using d to group, we simply type:
# Add means of 'a', grouped by 'd'
our_dt[, mean_a_by_d := mean(a), by = d]Finally, data.table as a useful function named uniqueN(), which is pretty much the combination of unique() and length()—it tells you the number of unique observations for whatever object you feed it.
How many unique values of d do we have?
# Count the unique values of 'd'
our_dt[,d] %>% uniqueN()## [1] 5Ordering
Last—also in the spirit of set() and setnames()—you can set the order of rows using setorder() and the order of columns using setcolorder().
Let’s order the rows of our_dt (arrange, in the dplyr world) by d and then by the reverse order (largest to smallest) of a:
# Order by 'd' and then reversed 'a'
setorder(our_dt, d, -a)
# Check work
our_dt## a b c d ab ac mean_a_by_d
## 1: 126 1.8050975 0.30503175 1 227.44229 38.434001 113.5
## 2: 121 0.7796219 0.87044788 1 94.33425 105.324194 113.5
## 3: 116 0.8168998 0.61894754 1 94.76038 71.797915 113.5
## 4: 111 -0.1162478 0.68835336 1 -12.90351 76.407223 113.5
## 5: 106 -1.8179560 0.21894784 1 -192.70333 23.208471 113.5
## ---
## 26: 125 -1.5977095 0.14451190 5 -199.71369 18.063988 117.5
## 27: 120 0.2987237 0.15009821 5 35.84684 18.011785 117.5
## 28: 115 -0.7505320 0.04825135 5 -86.31118 5.548906 117.5
## 29: 110 -0.9193220 0.94643075 5 -101.12542 104.107383 117.5
## 30: 105 0.6058875 0.97977825 5 63.61818 102.876716 117.5Again, there is no <- re-definition—the function does all of the work.
For ordering columns, you give setcolorder() the name of the data table and a character vector (or vector of integers for indexes) of the column names in your desired order. Let’s re-arrange our columns arbitrarily.
# Change column order
setcolorder(our_dt,
c("a", "ab", "ac", "mean_a_by_d", "b", "c", "d"))
# Check work
our_dt## a ab ac mean_a_by_d b c d
## 1: 126 227.44229 38.434001 113.5 1.8050975 0.30503175 1
## 2: 121 94.33425 105.324194 113.5 0.7796219 0.87044788 1
## 3: 116 94.76038 71.797915 113.5 0.8168998 0.61894754 1
## 4: 111 -12.90351 76.407223 113.5 -0.1162478 0.68835336 1
## 5: 106 -192.70333 23.208471 113.5 -1.8179560 0.21894784 1
## ---
## 26: 125 -199.71369 18.063988 117.5 -1.5977095 0.14451190 5
## 27: 120 35.84684 18.011785 117.5 0.2987237 0.15009821 5
## 28: 115 -86.31118 5.548906 117.5 -0.7505320 0.04825135 5
## 29: 110 -101.12542 104.107383 117.5 -0.9193220 0.94643075 5
## 30: 105 63.61818 102.876716 117.5 0.6058875 0.97977825 5Note that if you forget names, the function does not work.
More?
We’re only scratching the surface of the data.table package. The package also includes really efficient joins/merges (see ?data.table::merge). The merge() function has some cool properties—left joins, right joins, cartesian products, etc.—and you are also able to run rolling joins (using a different notation), which are awesome. Want more? You can add leads and lags easily with shift(), and the package includes fread() function for fast reading (though I think readr is a bit more stable and at least as fast, in general). And there are still more features.
data.table provides a number of great resources for learning:
I’m also happy to chat if you have questions, and I like coffee.5
Other nice tools for R
Here are a few more nice tools for R/research.
- New documentation website for
ggplot2! - A nicer way to read/search R’s documentation files.
- For administrative boundary shapefiles—includes
.rdsformat for reading straight into R!
Fun tools: SelectorGadget and rvest
rvest provides a simple and powerful package for webscraping in R. SelectorGadget provides an even more simple—and yet tremendously helpful—browser extension for finding CSS and xpath selectors. They make a great team.
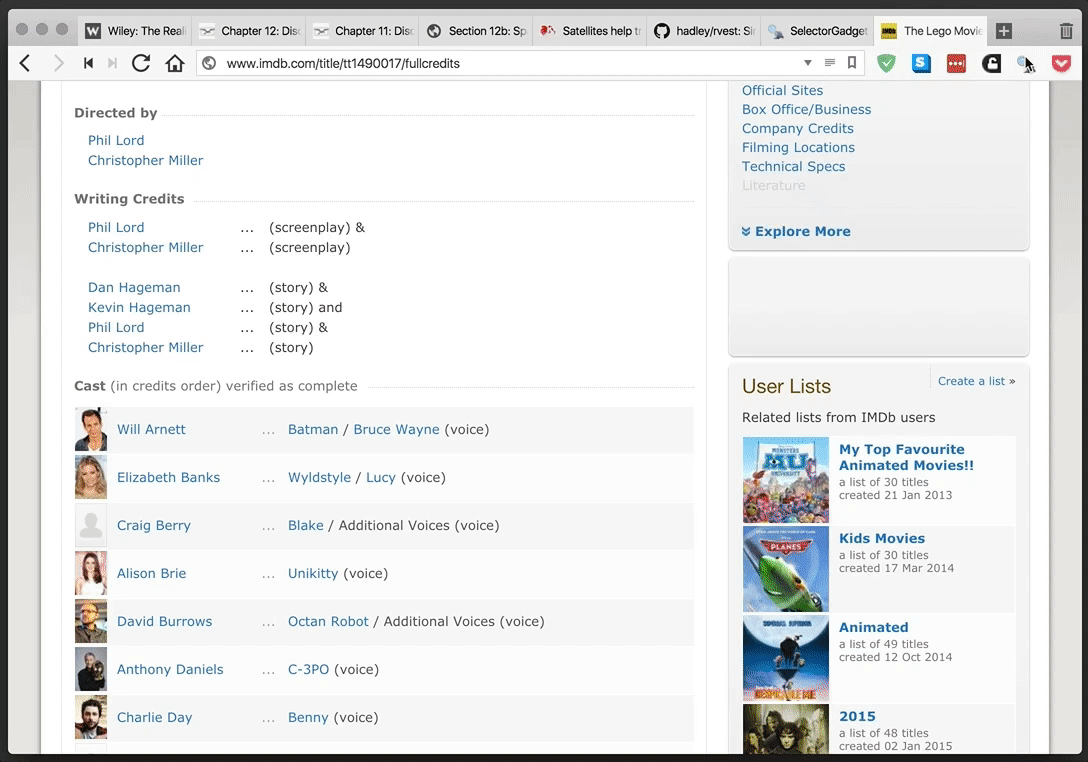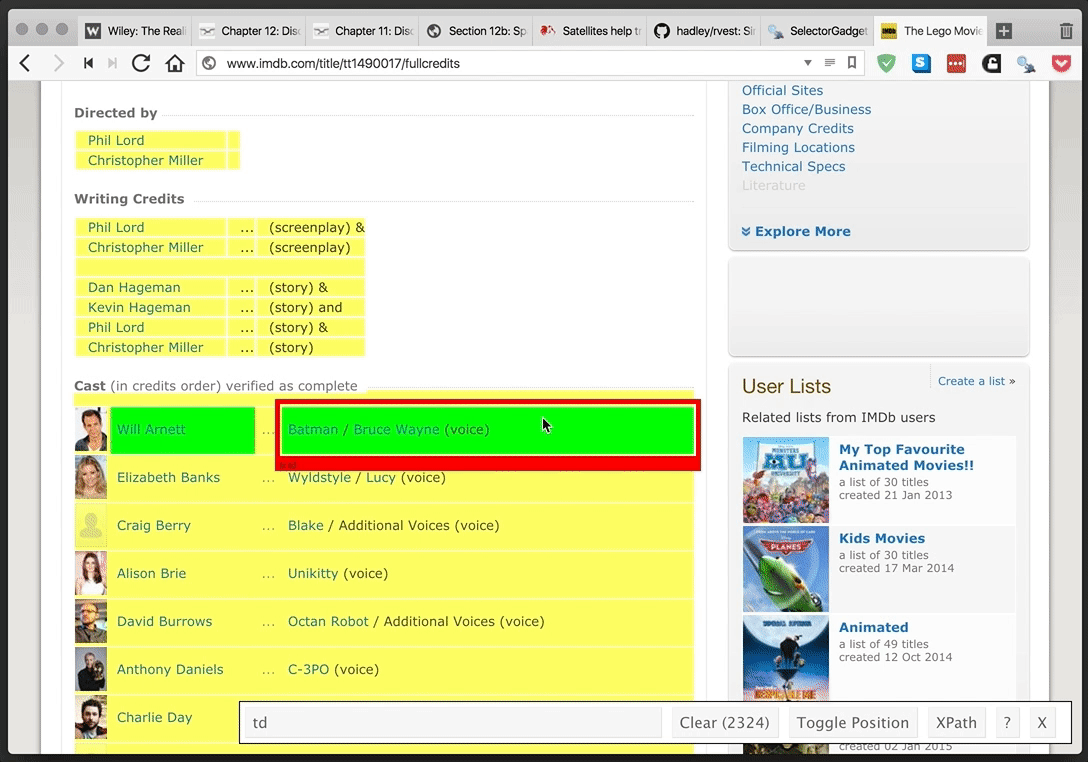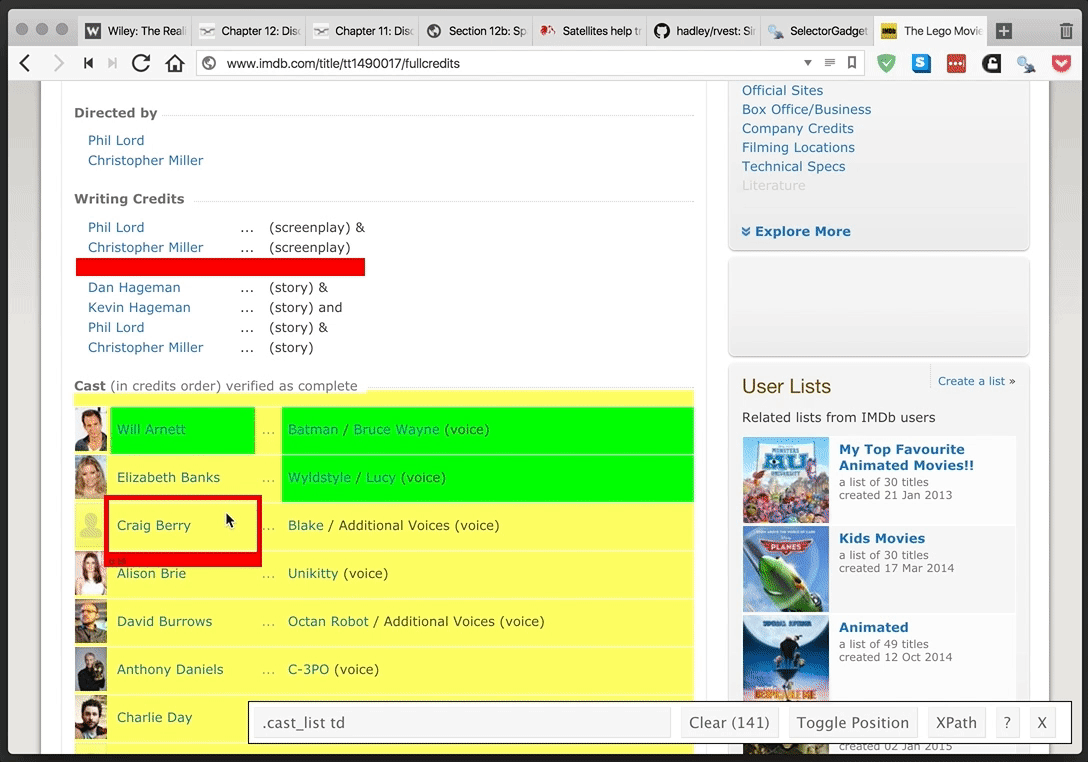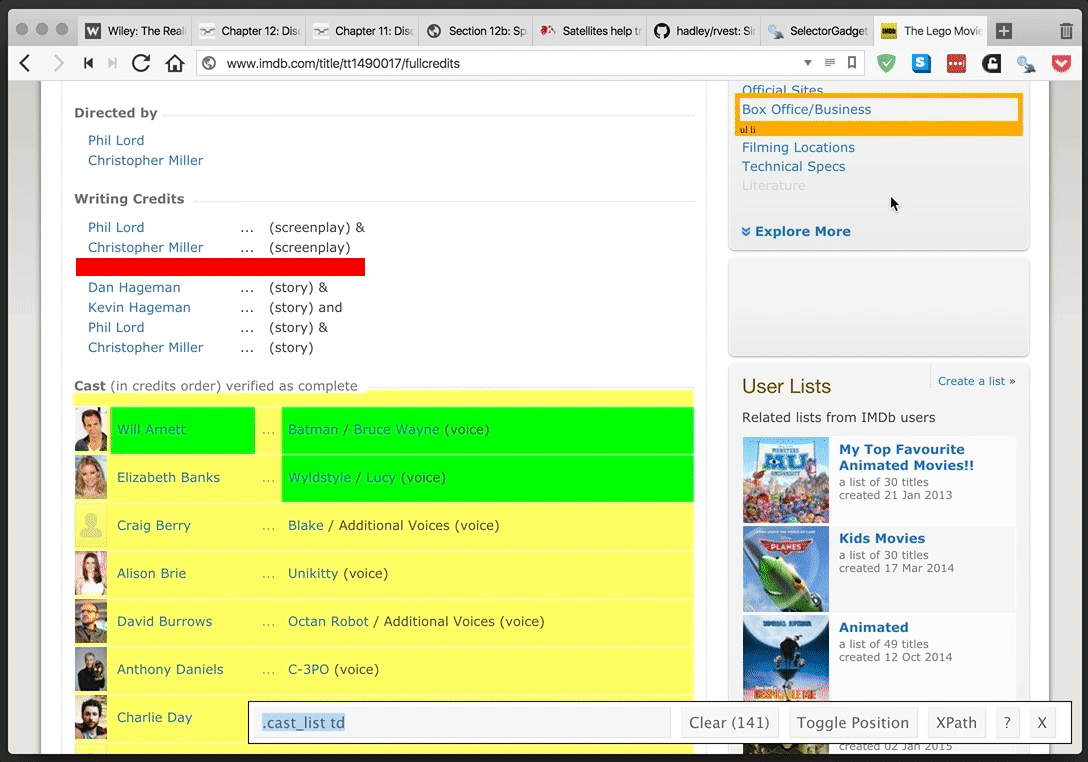 SelectorGadget in action: selecting the table of cast members for The Lego Movie from IMDB.
SelectorGadget in action: selecting the table of cast members for The Lego Movie from IMDB.
Bonus feature: Vivalid is a vastly customizable browser providing tab management and a host of other features not common to many other browsers.
tbl_dfs have a similar feature. Plus, R will not actually let you print a billion rows to screen, but it will allow you to print 50,000, which is still a lot.↩I’m going to write it “data table” rather than “data.table”, since written English is a bit different than scripting/programming languages.↩
Sampling? “Sub-sampling” seems redundant.↩
You could also stick with
$, but, again, it is inefficient. If you are switching todata.tablefor its efficiency/speed, it probably makes sense to avoid inefficient uses.↩Wink, wink.↩